Modern proteins use a nearly universal repertoire of 20 canonical amino acids, according to a team of researchers from the Charles University Faculty of Science, BIOCEV and IOCB (Czech Republic), Johns Hopkins University (U.S.), and ELSI (Japan) (AAs). The research, which was published in the Journal of the American Chemical Society, indicates that the canonical alphabet’s foldability was probably a key consideration.
Prior research indicates that only 10 “early” AAs were used by ancient proteins, and that the remaining 10 “late” AAs were byproducts of biosynthetic processes. Yet, a large number of non-proteinogenic amino acids (AAs) were prebiotically accessible, raising the questions of why we have the contemporary amino acid alphabet and whether proteins would be able to fold into globular forms if various amino acids made up the genetic code.
The team used experimental evaluation to determine the solubility and secondary structure propensities of a number of prebiotically significant amino acids in the context of synthetic combinatorial 25-mer peptide libraries to provide an answer to this question. To explore these alternate sequence spaces, the most prebiotically prevalent linear aliphatic and basic residues were added to or substituted for other early amino acids.
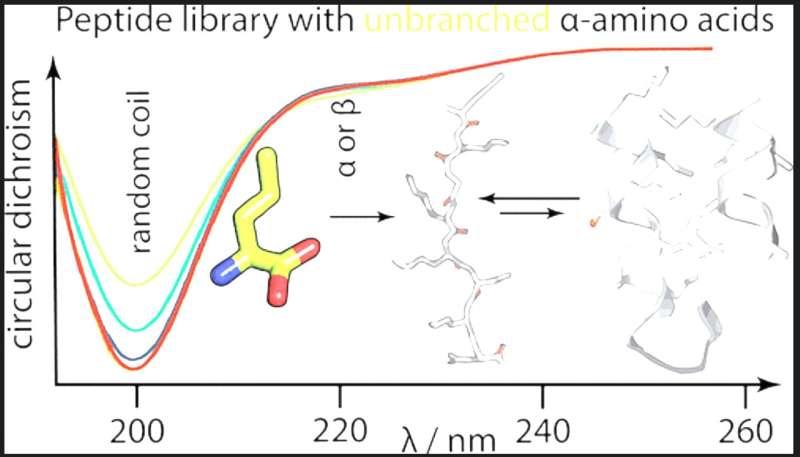
The findings demonstrate that despite their high prebiotic abundance, unbranched aliphatic amino acids were eliminated from the proteinogenic alphabet because they produce polypeptides that are excessively solubilized and have poor packing efficiency. Interestingly, the team proposes a biophysical explanation for how the presence of a short-chain basic amino acid reduces polypeptides’ secondary structure potential.
The results of the study lend support to the idea that, although missing basic residues, the early canonical alphabet was extraordinarily adaptable at facilitating protein folding and help to explain why basic residues were only added to proteins at a later stage of their evolutionary history.
Stephen D. Fried and Klara Hlouchova, the study’s primary authors, declared that they were “happy to have identified some of the reasons why the protein alphabet has developed in the way we know it currently.” Our findings demonstrate that foldability was a significant factor in the choice of the canonical alphabet and provide an explanation for the exclusion of some prebiotically accessible amino acids from the proteinogenic alphabet.
The results of the study provide light on the evolution of proteins and could have an impact on medication development and protein engineering. To examine the ramifications of their findings, the team intends to carry out additional investigation.
ALSO READ THIS: Using Clay’s Full Potential: Is It The Secret To Removing Carbon Dioxide From The Air?

